Tuberculosis (TB) is a communicable disease caused by an infectious bacterial pathogen called Mycobacterium tuberculosis (Mtb), which typically replicates in the lungs (pulmonary) but can affect other areas too. According to the World Health Organization (WHO), one third of the world’s population is infected with Mtb and 10 million people developed TB in 2018 with 1.4 million deaths globally. Historically, TB was considered incurable until the discovery and use of streptomycin (STR) in 1946. There has been a slow but good progress in the field of TB drug discovery and development.
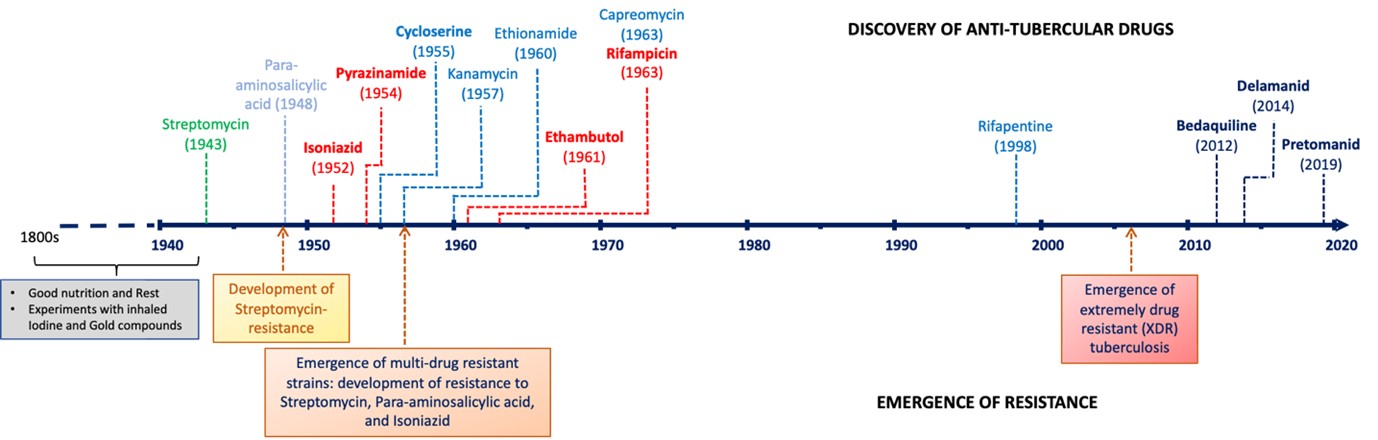
A timeline of TB drug discovery and emergence of resistance
In our research group, TB has been recognized as one of priority diseases for drug research and development. The expanding threat of multidrug-resistant tuberculosis (MDR-TB), co-infection with HIV, existence of latent TB infection and the extensive treatment regimen has prompted urgent calls for new approaches to TB control, in line with the global requirements, our research in the development of anti-TB drugs aims to:
(i) to discover/develop new chemical matter with novel mechanism(s) of action (MoA) – to treat drug-resistant strains and contribute to treatment shortening,
(ii) to find novel bactericidal compounds effective against persistent/dormant bacilli – to contribute to treatment-shortening, and
(iii) discovering drugs with no drug-drug interactions – for better drug-combinations.
To identify new chemical starting points for drug discovery, the target-based screening approach is preferred when target validation has been demonstrated. Selecting the target mainly relies on its vulnerability and selectivity. Interestingly, despite tremendous success in other disease areas, this approach was largely not successful in the TB drug discovery due to compound permeability issues across the lipid-rich cell-wall, efflux, and metabolization of the compound in Mtb. On the other hand, the phenotypic whole cell screening approach has relatively been more successful in delivering cell-permeable small molecules active against whole Mtb cells as starting points for TB drug discovery. This is then typically followed up by deconvolution of the MoA of the actives. Our programme has largely utilized this approach with small molecule active hits being progressed through the drug discovery process, underpinned by medicinal chemistry optimization, towards improving potency, pharmacokinetic, and pharmacodynamic properties. A critical benchmark in selecting a “New Chemical Entity” from the screening campaign is the representation of a new drug class, acting via a novel MoA. To achieve this, we typically perform hit-triaging at an early stage to avoid rediscovery of the established targets by using a slew of secondary assays. Our whole-cell based phenotypic screening assesses the growth kinetics by measuring the minimum inhibitory concentration (MIC) as the endpoint.
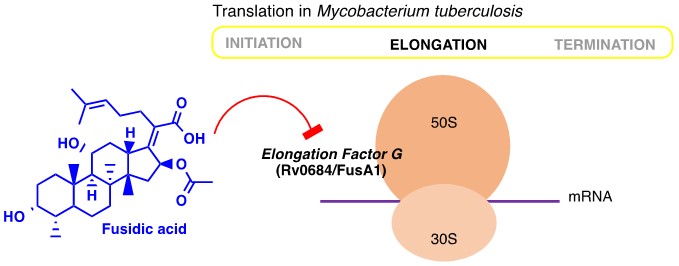
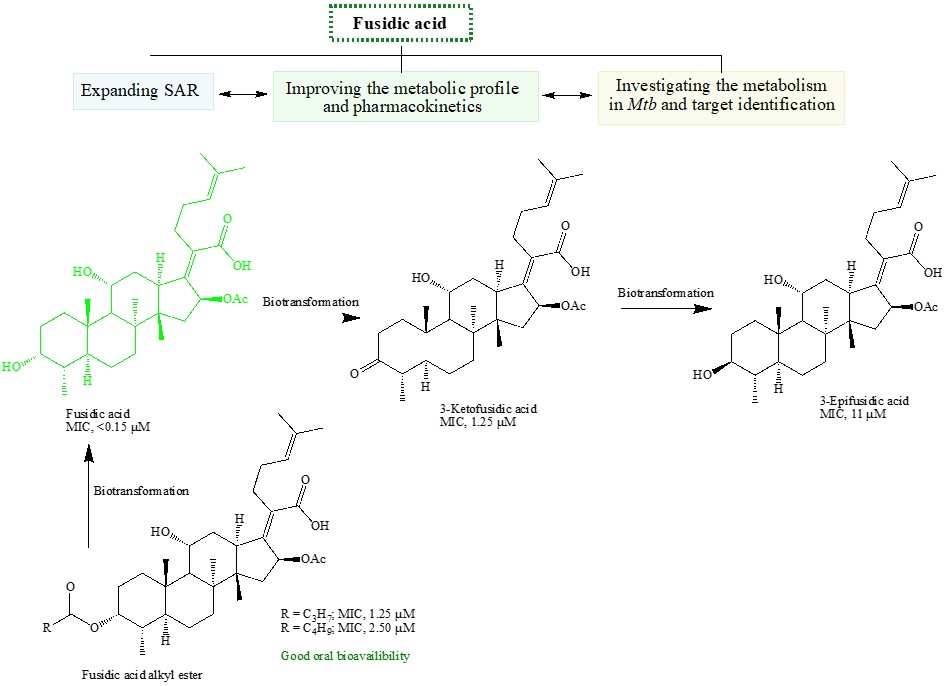
Additionally, one of the strategies in tackling drug-resistance is to reposition or repurpose existing drugs which can potentially reduce the cost and time of drug development. Towards this end, we investigated an experimental drug chlorpromazine (CPZ) and triterpenoid antibiotic fusidic acid (FA). Our current efforts are focussing on repurposing anti-cancer compounds as antibiotics.
Selected Publications:
DOI: 10.1021/acs.accounts.0c00878
DOI: 10.1021/acsinfecdis.1c00195
DOI: 10.1021/acs.jmedchem.1c00707
DOI: 10.1021/acsinfecdis.0c00252
DOI: 10.1016/j.drudis.2020.02.003
DOI: 10.1021/acsinfecdis.9b00208
More details of our TB research can be found in our publications [https://health.uct.ac.za/kelly-chibale-research/2020-2]
